The Road to Cybernaut-1: Rebuilding Search for AI
AI needs its own search engine. This is how we’re rebuilding search for AI -- and the road to Cybernaut-1, the first high-trust agentic search engine.

In the near future all searches will be done by AIs operating on our behalf. The problem is that search was not built for them. It was built for us - no, let's be honest - it was built for advertisers. The consequence is that AI agents are being kneecapped. We will fix that problem. We are Search, rebuilt for AI.
In this post, I will take you through our eight-stage multilingual retrieval pipeline step-by-step. I will touch on a variety of topics including tokenization, lexical search, semantic search, rank fusion, and more. Finally, I will share an important company update regarding an upcoming algorithm - cybernaut-1.
AI needs its own search engine
Building systems involves making trade-offs. How those trade-offs are decided cannot be disentangled from who your end user is. AIs are not people. People are not AIs. So, a search engine built for people will necessarily make different trade-offs to a search engine that is built for AIs. Take for example:
- Spell Checking: AIs don’t need spell-checking. "Did you mean" CPU cycles are better spent on detecting threats like prompt injections.
- Query Length: Human queries are short and vague. AI queries are long and unambiguous, which negates the need for personalization.
- Few-Shot Search: AIs can effortlessly generate multiple queries, so search engines that support multi-query input are more valuable.
- Recall Maxxing: People need precision; AIs need recall. Short attention spans (people) versus massive context windows (AIs).
- Complexity: Human search engines must be idiot-proof. AI search engines must be the opposite: genius-friendly. Why? Because a future where every search is done by AI is a future where every search is done by a SQL expert who speaks 200 languages and knows everything.
I could go on for days about this, but you get the point: search engines are not aligned with AIs and AIs are paying the price. So, we are building the world's most AI-aligned search engine. Let's dive into how it actually works.
Today we are 250,000 search engines
From the outside, NOSIBLE looks and behaves like one large search engine. On the inside, NOSIBLE is actually made up of a federation of 250,000 smaller search engines called shards. Each shard acts like a giant magnet, attracting similar texts. Collectively they house almost a billion webpages, billions of embeddings, and hundreds of billions of words of text.
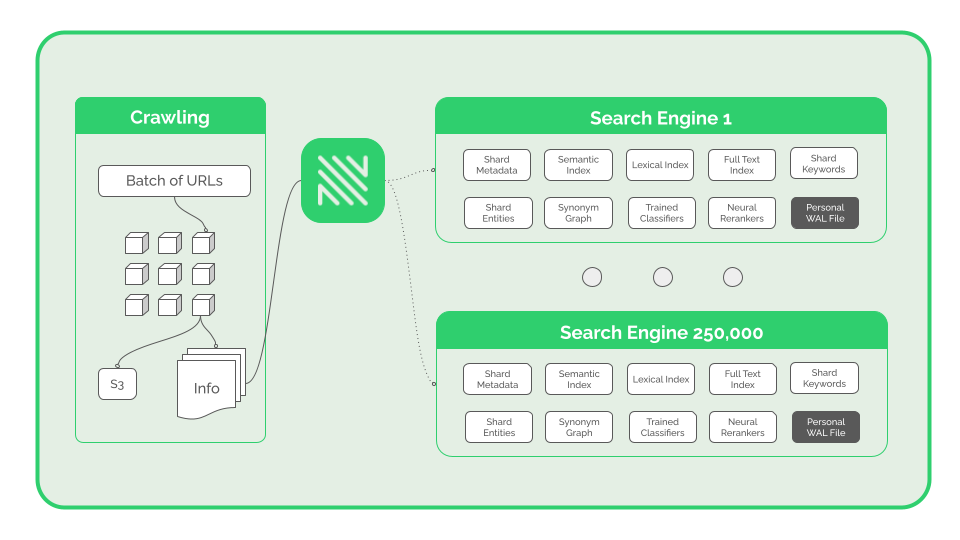
If you opened up a shard you would find:
- Metadata - Descriptions, classifications, and more.
- Full-Text Index - Used for longer n-gram retrieval.
- Lexical Index (BM25) - Used for keyword-based retrieval.
- Semantic Index - Used for vector-based retrieval.
- Vocabulary - The set of unique words in this shard.
- Keywords - Important words to this shard.
- Entities - Important entities to this shard.
- Synonyms - A graph learned from word co-occurrences.
- Classifiers - Fine-tuned classifiers for signals e.g. sentiment.
- Compressors - Trained compression dictionaries.
- Bloom Filters - Used to quickly check if phrases exist.
- Queries - Examples of queries that align with this shard.
- Evals - Gold standard datasets for evaluating this shard.
- Rerankers - Fine-tuned neural reranking models.
- Write-Ahead Log - Pending entries to the shard.
And hopefully soon you would also find:
- LoRAs - Shard Low-Rank Adaptations.
- Small LLMs - 0.5-2Bn parameter LLMs (more later).
Each shard is independently capable of performing lexical retrieval, semantic retrieval, full-text retrieval, hybrid retrieval, and more complex tasks.
Each shard can be deployed on its own or alongside others. This gives us a lot of options for scaling up to 10+ billion webpages within the next 6 months.
Shards are learned using an efficient algorithm we developed for identifying semantically and lexically coherent clusters of texts that are also approximately uniformly sized. Uniformity helps ensure that queries per second per shard is predictable. Our most recent training run involved 250,000,000 embeddings. Our next run will involve 1 billion embeddings and a lot of GPU time.
Let's take shard 11,343 as an example:
11,343: CRISPR Genome Editing Using sgRNA, Plasmids, Nucleases, and Homologous Recombination in Bacterial and Yeast Systems
11,343 focuses on CRISPR-based genome editing methods involving sgRNA, crRNA, and nucleases to induce targeted DNA cleavage. It covers plasmid design, transfection, and the use of homologous recombination and non-homologous end joining for mutation and gene insertion. The content includes molecular tools like PCR, cloning, and protein expression in bacterial and yeast models. It addresses spacer sequences, codon optimization, and biosynthetic pathways, highlighting applications in mutagenesis, transgenesis, and metabolic engineering. Key elements include DNA repair mechanisms, vector construction, and functional assays to study gene function and phenotype changes.
- Classifications:
- Language: Mostly English
- Geography: Worldwide
- Brand Safety Classification: Safe
- Industry Classification: Biotechnology
- IAB: Biotech and Biomedical Industry
- Scope & Size:
- Number of documents: 134,523
- Number of words: 8,446,510
- Vocabulary size: 64,992
- Number of keywords: 2,523
- Top 20 Keywords:
- plasmid → plasmid, plasmids
- crispr → CRISPR, CRISPR-Cas9, CRISPR-Cas systems
- sgrna → single guide RNA, sgRNA
- genom → genome, genomic, genomes
- grna → guide RNA, gRNA
- rna → RNA
- nucleas → nuclease, nucleases (e.g., Cas9 nuclease)
- crrna → CRISPR RNA, crRNA
- sgrnas → single guide RNAs, sgRNAs
- transfect → transfect, transfection, transfected
- nucleotid → nucleotide, nucleotides
- grnas → guide RNAs, gRNAs
- streptomyc → Streptomyces (bacterial genus often used in biotechnology)
- rnas → RNAs (plural of RNA, could be mRNAs, tRNAs, etc.)
- spacer → spacer sequence(s), CRISPR spacer
- codon → codon, codons, codon optimization
- peptid → peptide, peptides
- strep → Streptococcus
- gfp → green fluorescent protein (GFP)
- recombin → recombination, recombinant, recombine
- Top 20 Entities
- CRISPR Therapeutics - Mentioned 6408 times
- Thermo Fisher Scientific Inc. - Mentioned 2027 times
- Addgene - Mentioned 1551 times
- Qiagen - Mentioned 1374 times
- Illumina - Mentioned 1204 times
- Invitrogen - Mentioned 1160 times
- New England Biolabs - Mentioned 1137 times
- Editas Medicine - Mentioned 1014 times
- British Gynaecological Cancer Society - Mentioned 938 times
- Intellia Therapeutics - Mentioned 884 times
- Life Technologies - Mentioned 800 times
- Pacific Biosciences - Mentioned 648 times
- Takara Bio - Mentioned 565 times
- Agilent Technologies - Mentioned 558 times
- Mammoth Biosciences - Mentioned 529 times
- Sigma-Aldrich - Mentioned 496 times
- Promega - Mentioned 437 times
- GenBank - Mentioned 421 times
- Integrated DNA Technologies - Mentioned 401 times
- Millipore Corporation - Mentioned 397 times
And that's just one! We have 250,000 of them and collectively they span every aspect of the human condition past and present. They also provide a unique glimpse into how models perceive / misperceive our world.
The journey from questions to answers
Every question that comes into NOSIBLE goes through a pretty sophisticated eight-stage retrieval pipeline. These eight stages are as follows:
- Language Detection and Translation
- Multilingual Tokenization
- Search Intent Prediction
- Instruction Tuning and Embedding
- Shard Selection
- Shard Reranking
- Shard-based Question Expansion
- Retrieval using Map-Reduce
Let's follow "What lessons from bacteria and yeast actually translate into safer gene-editing medicines?" through the pipeline in English and in Japanese.
1 // Language Detection and Translation
Due to the ever shrinking - but unfortunately still present - language gap in multilingual embedding models it is still necessary to detect what language the question and expansions were written in and, if results are requested in a different language, translate that question into the requested language.
Worked Example
- "What lessons from bacteria and yeast actually translate into safer gene-editing medicines?" → "What lessons from bacteria and yeast actually translate into safer gene-editing medicines?" (no translation).
- "What lessons from bacteria and yeast actually translate into safer gene-editing medicines?" → "細菌と酵母から得られる教訓は、より安全な遺伝子編集医薬品にどのように応用されるのでしょうか?".
Tech Stack
- fasttext-langdetect - supports 170+ languages.
- gemma-3n-e4b-it - supports 140 languages.
2 // Multilingual Tokenization
Next, the question and expansions are tokenized. This involves sentence boundary detection, text segmentation, text inflection, Unicode normalization, word stemming, stop-word removal, and other classic NLP techniques.
Getting this to work equally well across many languages is difficult. We ended up creating a standardized interface that wraps probably a dozen or more different NLP packages and abstracts away all language-specific complexities.
I know it might sound strange to be working on this in 2025. Why not just use an LLM? Why not just do semantic search? The answers to those questions are (1) even small LLMs are too slow and (2) semantic search is not a silver bullet.
In all honesty, well-calibrated lexical search indices beat well-calibrated semantic search indices on AI queries more often than not. Why? Because AI queries are longer and have high intent. That said, hybrid search dominates both.
Worked Example
Q: "What lessons from bacteria and yeast actually translate into safer gene-editing medicines?" yields the following tokens:
- "yeast"
- "bacteria"
- "safer"
- "lesson"
- "translat"
- "gene"
- "medicin"
- "edit"
- "actual"
Q: "細菌と酵母から得られる教訓は、より安全な遺伝子編集医薬品にどのように応用されるのでしょうか?" yields the following tokens:
- "細菌" (bacteria)
- "教訓" (lesson)
- "遺伝子" (gene)
- "酵母" (yeast)
- "応用" (application)
- "治療" (treatment)
- "編集" (edit)
- "どの" (which)
- "安全" (safety)
Tech Stack
- SpaCy - full/partial support for 75 languages
- NLTK - mixed support for ~50 languages.
- BlingFire - English only (super-fast)
- pySBD - support for 22 languages
- pyStemmer - support for 24 languages
- indic-NLP-library - supports Indian languages
- pymorphy3 - support for Russian and Ukrainian.
- jieba - support for Chinese text.
- mecab-python3 - support for Japanese text.
- mecab-ko - support for Korean text.
- wtpsplit - 85 languages (incredibly slow).
- ipadic - support for Japanese text.
- inflect - support for English text only.
- regex - sigh. Way too much regex.
3 // Search Intent Prediction
Next, we run the tokens and their TF-IDF scores through a system that identifies search intents. We define a search intent as a sequence of proximal tokens that have a high "harmonic TF-IDF score" and do not contain certain parts-of-speech like, for example, pronouns, determiners, conjunctions, adverbs, etc.
Worked Example
Q: "What lessons from bacteria and yeast actually translate into safer gene-editing medicines?" yields the following search intents:
- "editing medicine"
- "gene-editing"
- "gene-editing medicine"
- "safer gene"
- "safer gene-editing"
- "safer gene-editing medicine"
Currently search intents are not available in Japanese. We expect to have search intents working for all supported languages in the week.
Tech Stack
4 // Instruction Tuning and Embedding
Next, we generate an appropriate instruction for the question and submit it, along with the expansions, to multilingual-e5-large-instruct. E5 is an open-source, instruction-tuned embedding model from Microsoft Research. It is designed to align vectors with natural language instructions.
A while back we used an LLM to generate and "evolve" optimal instruction templates for E5. Here is one of the most successful templates:
"Given a question, please retrieve any relevant {Language} Headlines, Leads, Passages, and Source URLs that focus on the same named entities as the question, and provide substantive answers to the question."
We have templates that have placeholders for named entities, geographic regions, topics of interest, and other dimensions. In our internal evals, we have seen that optimizing the instruction - or getting a small LLM to write a bespoke one - can yield a free 1-5% improvement in search precision and recall.
Tech Stack
5 // Shard Selection
Next, we move on to selecting the best shards for the question. This is arguably the single most important stage in our retrieval pipeline because, if we route the question to the wrong shards, we won't return the best document. Our current approach involves computing four ranking factors, namely:
- Vanilla Dense Similarity - This selector computes the cosine similarity between the E5 embedding of the question and E5 embeddings of the LLM-generated summaries for each shard (uses HNSW).
- Bayesian Dense Similarity - This selector is derived from a vector search index we developed that often beats the current state-of-the-art. I can't discuss it because we are pursuing a patent on it 😉.
- Vanilla Sparse Similarity - This selector computes the sparse similarity between the keywords in the question and the keywords in each shard. It's essentially just a massive sparse TF-IDF matrix.
- Entity Sparse Similarity - If the question contains one or more entities, we also compute the sparse similarity between those entities and the entities in each shard. Again, it is a massive sparse matrix.
These ranking factors are then combined using reciprocal rank fusion (RRF). RRF is a simple but powerful ensembling method that merges multiple ranked lists by giving higher weight to items that appear near the top across rankings. This gives robust results even if one of the ranking factors is noisy.
Because our shards are trained to be maximally semantically and lexically coherent, we are able to select the best shards almost all of the time. The only time we struggle is for very short queries. But, like I said, we are building NOSIBLE for AIs, not people, so this is a trade-off we are happy to make.
Worked Example
Q: "What lessons from bacteria and yeast actually translate into safer gene-editing medicines?" is routed to these English shards:
- 220,067
- Title: CRISPR Genome Editing RNA Enzymes Plasmids Epigenetics Molecular Biology Protein Mutation Recombinant Technologies
- 49,619
- Title: Overview of CRISPR Genome Editing Technologies and Molecular Mechanisms in Genetic Engineering and Therapeutics
- 178,396
- Title: Overview of GMO Safety, Toxicology, Biotechnology, Regulatory Agencies, and Food Additives Impacting Agriculture and Health
- +97 others
Q: "細菌と酵母から得られる教訓は、より安全な遺伝子編集医薬品にどのように応用されるのでしょうか?" is routed to these Japanese shards:
- 199,016
- Title: 量子コンピューティング技術開発と応用に関する研究とシステム設計の最新動向と課題解決方法
- 220,938
- Title: セキュリティとサイバーセキュリティに関するデータ保護システム開発と企業向け安全管理ソリューションの概要
- 111,375
- Title: 企業のデジタルサービス開発とプラットフォーム活用によるビジネス成長と技術課題解決の方法論
- +97 others
Tech Stack
6 // Shard Reranking
In the previous step the shards we selected for the Japanese question were quite bad. The first shard is about quantum computing, the second shard is about data protection, and the third shard is about digital platforms. This is where shard reranking comes in. Reranking involves reordering the selected shards by estimating their actual relevance to the question, so that the best shards rise to the top and irrelevant ones (like the ones we selected) get pushed down.
We have quite a few rerankers:
- Bloom Filter Reranker - Each shard has a bloom filter that keeps track of important phrases it contains. If a selected shard hasn't seen any of the user's search intents, then that shard should get downranked.
- Compression Reranker - Each shard has a trained Zstandard text compression dictionary. The shard that can compress the input question the best is more likely to contain the most similar content.
- Neural Reranker - We can also feed the LLM-generated titles and descriptions of each selected shard along with the question into a neural reranker like bge-reranker-v2-m3 to get a reranked set of shards.
- Page (Re)Ranker - Last, but certainly not least, it is also possible to use a nearest neighbor graph and the Personalized PageRank algorithm to produce a probability mass over your selected shards.
Once again, these ranking factors are then combined using reciprocal rank fusion (RRF). We have also used LLMs to rerank selected shards. That works very well but the added latency is simply too high for a search engine.
Worked Example
In the case of the Japanese shards the one and only shard we have that talks about gene editing in Japanese bubbles to the top. In the case of the English shards the shard that actually talks about bacteria and yeast in the context of gene editing bubbles up to the third position. Which is what we want to see.
Q: "What lessons from bacteria and yeast actually translate into safer gene-editing medicines?" is routed to these English shards:
- 49,619
- Title: Overview of CRISPR Genome Editing Technologies and Molecular Mechanisms in Genetic Engineering and Therapeutics
- 220,067
- Title: CRISPR Genome Editing RNA Enzymes Plasmids Epigenetics Molecular Biology Protein Mutation Recombinant Technologies
- 11,343
- Title: CRISPR Genome Editing Techniques Using sgRNA, Plasmids, Nucleases, and Homologous Recombination in Bacterial and Yeast Systems
- +97 others
Q: "細菌と酵母から得られる教訓は、より安全な遺伝子編集医薬品にどのように応用されるのでしょうか?" is routed to these Japanese shards:
- 200,739
- Title: 医薬品開発と臨床試験における製薬企業の治療法承認市場展開と感染症ワクチン技術動向分析
- 30,173
- Title: 睡眠の質向上に関する研究と健康影響 食事習慣 ホルモンバランス ストレス対策 運動効果 栄養摂取の重要性
- 168,764
- Title: ストレス管理と健康維持に関する食事、栄養素、メンタルヘルス、育児、免疫、生活習慣の重要性について
- +97 others
The translation of shard 200,739 in English is "Analysis of Pharmaceutical Companies’ Therapeutic Approval, Market Deployment, and Infectious Disease Vaccine Technology Trends in Drug Development and Clinical Trials".
Tech Stack
7 // Shard-based Question Expansion
Next, we use the synonym graphs in each shard to probabilistically expand our search terms. Because each shard is lexically and semantically coherent, the synonyms in each shard are unambiguous. Put simply, "gene" in shard 11,343 only has the genetic meaning. We don't need to worry that Gene is also a name and, in some other shards, would co-occur a lot with "Willy Wonka".
It's worth mentioning that this step can introduce some noise / serendipity, so we are very careful not to overwhelm the original search words.
Worked Example
Q: "What lessons from bacteria and yeast actually translate into safer gene-editing medicines?" when expanded produces the following:
- Original Search Words:
- "yeast"
- "bacteria"
- "safer"
- "lesson"
- "translat"
- "gene"
- "medicin"
- "edit"
- "actual"
- Expanded Search Words
- "yeast"
- "bacteria"
- "safer"
- "lesson"
- "translat"
- "gene"
- "medicin"
- "edit"
- "actual"
- "invad" [NEW]
- "palindrom" [NEW]
- "bacteriophag" [NEW]
- "interspac" [NEW]
- "antibiot" [NEW]
- "archaea" [NEW]
- "bacterium" [NEW]
- "pathogen" [NEW]
- "phage" [NEW]
- "dextros" [NEW]
- "infecti" [NEW]
- "raffinos" [NEW]
- "biofuel" [NEW]
- "galactos" [NEW]
- "acronym" [NEW]
- "chromosom" [NEW]
- "crispr" [NEW]
- "insect" [NEW]
- "prokaryot" [NEW]
- "mojica" [NEW]
- "bacto" [NEW]
Currently question expansions are not available in Japanese. We expect to have question expansions working for all supported languages in the week.
Tech Stack
8 // Retrieval Using Map-Reduce
At this point we finally have (1) our instruction-optimized embeddings, (2) our expanded set of search terms, (3) our search intents, and (4) a list of 10-30 shards that almost certainly contain documents that relate directly to our question. This package of information is then broadcast to each of the final shards.
When received each shard will:
- Run the user's SQL filter to filter out documents.
- Execute a full lexical search over the valid records.
- Execute a full semantic search over the valid records.
- Fuse the lexical and semantic results together.
- Then do a full-text search over top results for intents.
The results from each shard are combined and grouped by document identifier. Once the search completes a highly relevant snippet is constructed for each search result. The maximum length of the snippet is decided by the user.
Worked Example
Q: "What lessons from bacteria and yeast actually translate into safer gene-editing medicines?" yields the following search results:
🔗 Yeast could prove game-changer in medicine
In this project, we use genes sourced from plants to make a metabolite that is produced by plants such as oranges — we're producing naringenin, a molecule produced by fruit trees like lemons, grapefruits or oranges," said Molecular microbiologist Jean-Marc Daran of the Delft University of Technology. To reprogram yeast, researchers at the Delft University of Technology are using an editing process called CRISPR. The procedure inserts genes from plants or bacteria into yeast. That can change how the cell factories work — and even how they smell! "We edited the DNA, and we've added among others one gene from a plant. And because of that it now also really smells like roses. Your lab smells better of course, but we are more looking into the industrial applications so we can smell this as a flavour and aroma compound — mostly for the cosmetic industry, mostly perfumes but also mascara, lipstick — they all have this flavour compound in them," metabolic engineering researcher Jasmijn Hassing told Euronews.
+99 more results
Q: "細菌と酵母から得られる教訓は、より安全な遺伝子編集医薬品にどのように応用されるのでしょうか?" yields the following search results:
🔗 あらゆる産業の常識を変えうる「エンジニアリングバイオロジー」が期待されるわけ
エンジニアリングバイオロジーとは、 バイオテクノロジーを用いて有用な機能を持った生物(スマートセル)を開発し、バイオプロセスによって有用物質を生産する技術領域 のことを指します。 スマートセルとは、バイオテクノロジーによって有用物質の生産能力を向上させた、細菌や酵母、植物などを総称した日本での呼び名です(海外では「Engineered organisms」や「Engineered cells」など)。 スマートセルを活用したバイオプロセスによるバイオものづくりは 医薬品、食料、燃料、衣類など幅広い分野への応用ができます 。
Engineering biology refers to a technological domain in which organisms with useful functions (“smart cells”) are developed using biotechnology, and useful substances are then produced through bioprocesses. The Japanese term “smart cells” collectively refers to bacteria, yeast, plants, and other organisms whose ability to produce useful substances has been enhanced through biotechnology (overseas, they are called things like “engineered organisms” or “engineered cells”). Bio-manufacturing based on smart cells and bioprocesses can be applied across a wide range of fields, including pharmaceuticals, food, fuel, and clothing.
+99 more results
Tech Stack
How does this tie into agentic search?
As I mentioned earlier - Human search engines must be idiot-proof; AI search engines must be the opposite: genius-friendly. In the coming days we will launch our V2 search API which includes an algorithm called cybernaut-1.
Cybernaut : A voyager in cyberspace, indicating someone who navigates or explores online environments.
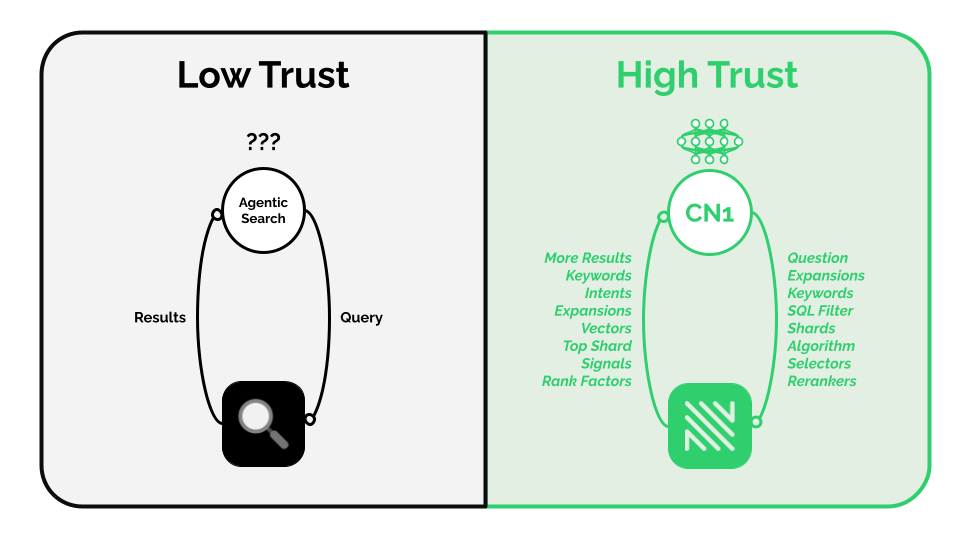
Cybernaut-1 is an AI agent with unrestricted access to everything in NOSIBLE including every shard, algorithm, selector, reranker, and signal. It knows what these things are and can tune them on the fly to find better results.
Other agentic searchers, by comparison, are flying blind. They only see the top search results. They don't know how they got there. So, their ability to reflect and iterate is fundamentally constrained. It's just a random walk.
Put differently, cybernaut-1 is a reinforcement learner that designs and iterates on search policies to maximize search relevancy. It is the culmination of months of work, and it will be generally available on the 25th of August 2025.
Epilogue.
At NOSIBLE we believe that search is a scaling law on par data or compute. Giving AIs access to 10x, 100x, 1000x more search will make them more intelligent and more capable. Recent research has hinted that this is true.
We believe this because it reflects how we work. Our intelligence is not an island. Nobody expects us to recall every fact from memory. We are deeply connected to knowledge 24/7. We seek it, read it, grok it, and act upon it.
AIs will do the same, only at superhuman scale. AIs will seek knowledge millions of times a day in ways that are markedly different from you or I. So, they need their own search engine. Search isn't a tool call - it is intelligence.

